|
Texture
In works Memo |
Multifit /
Extraction Of Chi Filesor how to slice up your diffraction images...General discussionFit2d will be used to obtain slices of the diffraction images as a function of azimuth. Typically, your image shows the Debye-Sherrer rings, slightly distorted because of the deviatoric stress applied to your sample. They run from a azimuth angle Typically, in deformation studies, you want to slice your data with a given interval (5 degrees), and, for each angle, have tabulated data with the intensity as a function of the diffraction angle Multifit assumes that your diffraction data was named with a base and indices, for instance test_0003.tif test_0004.tif test_0005.tif .... test_0236.tif where You will want to slice up all those images, typically, every 5 degrees for test_0003_0.chi test_0003_5.chi test_0003_10.chi ... test_0003_355.chi test_0003_360.chi test_0004_0.chi ... test_0004_360.chi ... test_0236_0.chi ... test_0236_360.chi Creating a test macroI do not recommend to go ahead creating the full macro that will create the thousands of chi files without some little testing: do not start with the full range! Do it for a couple of images first (0003 to 0010 in our example, for instance). Make sure everything on this page works, and then create the full macro. Otherwise, you might be overheating your processor for 2 to 3 hours for nothing... To create your macro, open 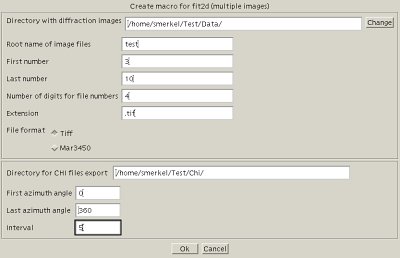 Create a macro for Fit2d In our case, if we want to slice images 3 to 10, every 5 degrees for azimuth between 0 and 360 degrees, we will enter
It will create a file called Running the macroIn fit2d, exit the powder diffraction interface, and select  Run a macro in Fit2d You should see fit2d moving quite a bit, slicing images and so on. In the Fit2d log file, you can follow what is going on (it might go quite fast). Try to make sure that images are loaded properly, and that chi files are also saved properly. For instance, our
/home/data/Test/test_0003.tif: Azimuth/2-theta
2-Theta Angle (Degrees)
Intensity
540
8.9983260E-03 0.0000000E+00
2.6994979E-02 0.0000000E+00
4.4991631E-02 0.0000000E+00
6.2988281E-02 3.1282605E+02
8.0984935E-02 3.1098578E+02
9.8981589E-02 3.0880267E+02
1.1697824E-01 3.1000055E+02
1.3497490E-01 3.0902777E+02
1.5297155E-01 3.0717679E+02
...
The first line is the full path to the image. If the full path is not shown, something is wrong!! If you see Second and third lines are labels. Fourth line is the number of points, later on, you have 2 theta in the first column and the intensity in the second column. |